|
Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures |
Tutorials
| Analysis | Visualization | Rebuilding | Composite | Fiber | Mutation |
The mutation component of Web 3DNA 2.0 allows the user to mutate any base(s) of a nucleic acid structure to desired one(s). The user can use a PDB/NDB ID, a user-uploaded PDB-formatted file, or any of the models constructed by the modeling components in the web server. The output includes: 1) schematic representations of the original and mutated structures, 2) a PDB-formatted file with full atomic coordinates of the mutated structure, and 3) 3D interactive visualizations of the original and mutated structures.
It should be noted that both the sugar-phosphate backbone conformation and base reference frame (position and orientation) are preserved in the mutation process. Therefore, the original and mutated structures share the same base-pair/step parameters.
Example 6-1: Mutate a nucleic-acid-containing structure by using the PDB/NDB ID
The user first enters a PDB ID (for example "355D") and clicks the 'List all bases' button.
Then the user is directed to an intermediate page where an enumeration of all bases is listed.
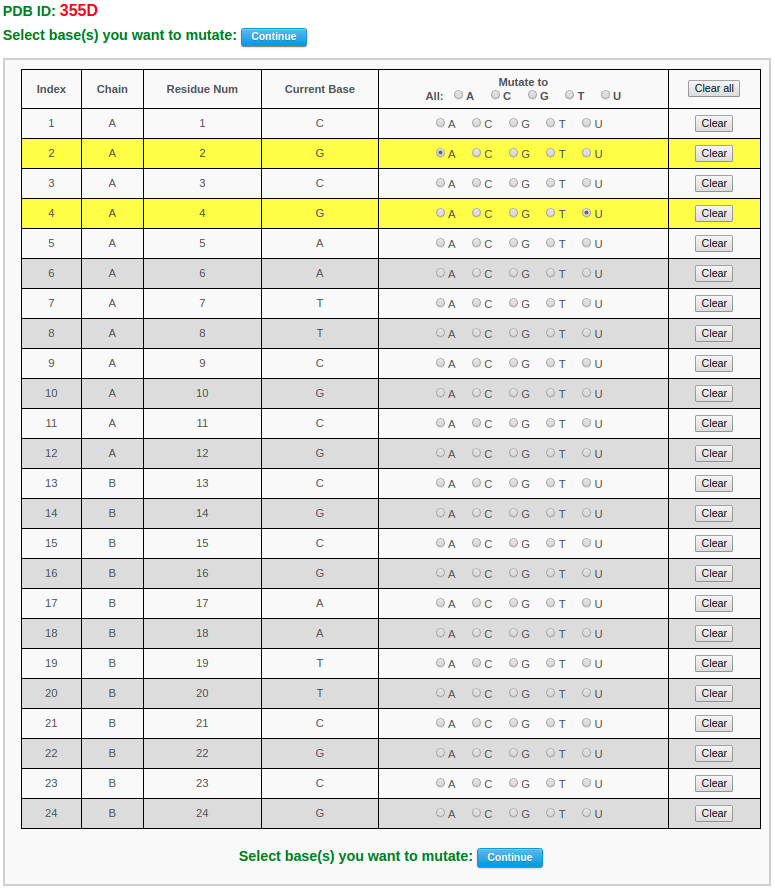
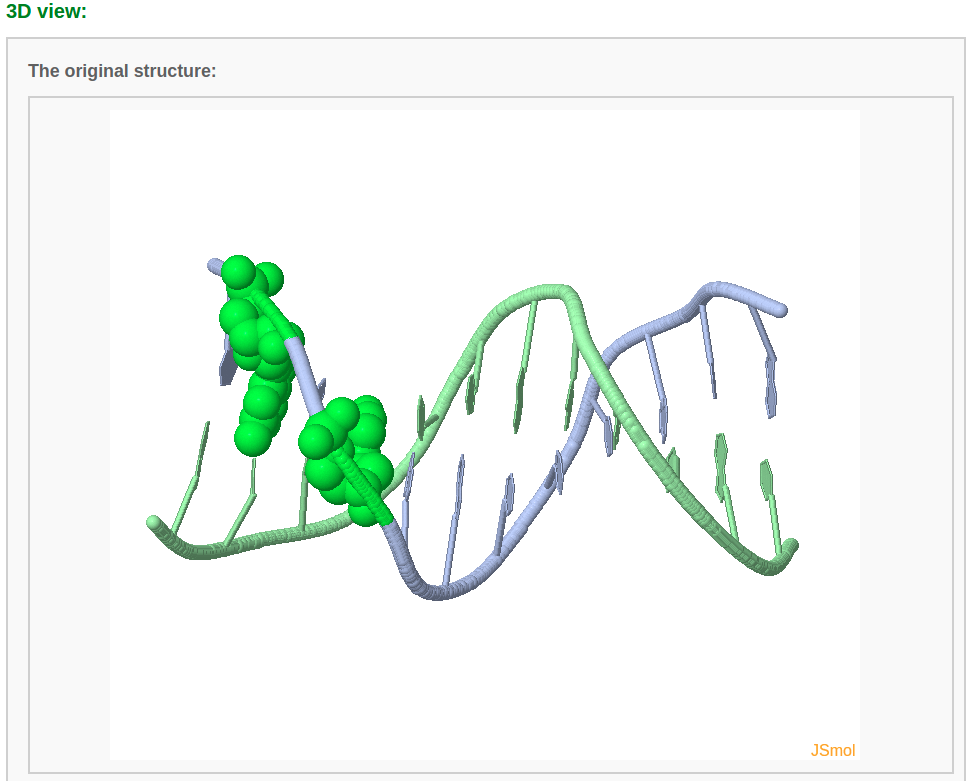
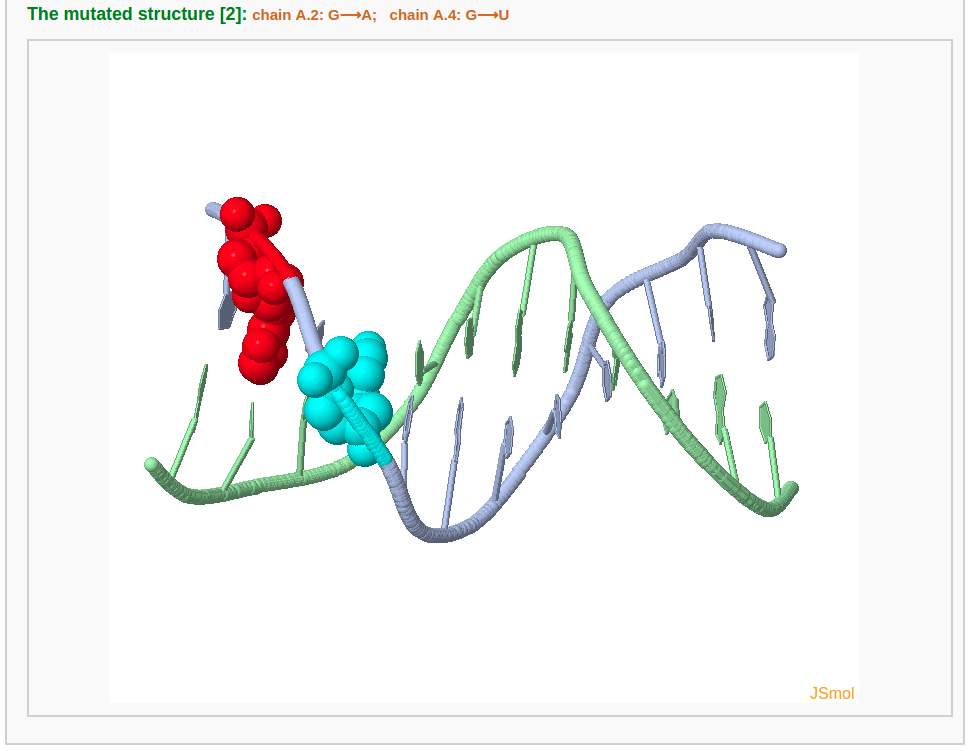
After choosing base(s) for mutation (for example here we mutate base G in chain A/residue number 2 to A and base G in chain A/residue number 4 to U) and clicking the "Continue" button, the user will be directed to a results page containing five sections: 1) a block representation of the original structure, 2) a list of mutated bases, 3) a block representation of the mutated structure, 4) a 3D interactive visualization of the original structure, 5) a 3D interactive visualization of the mutated structure.
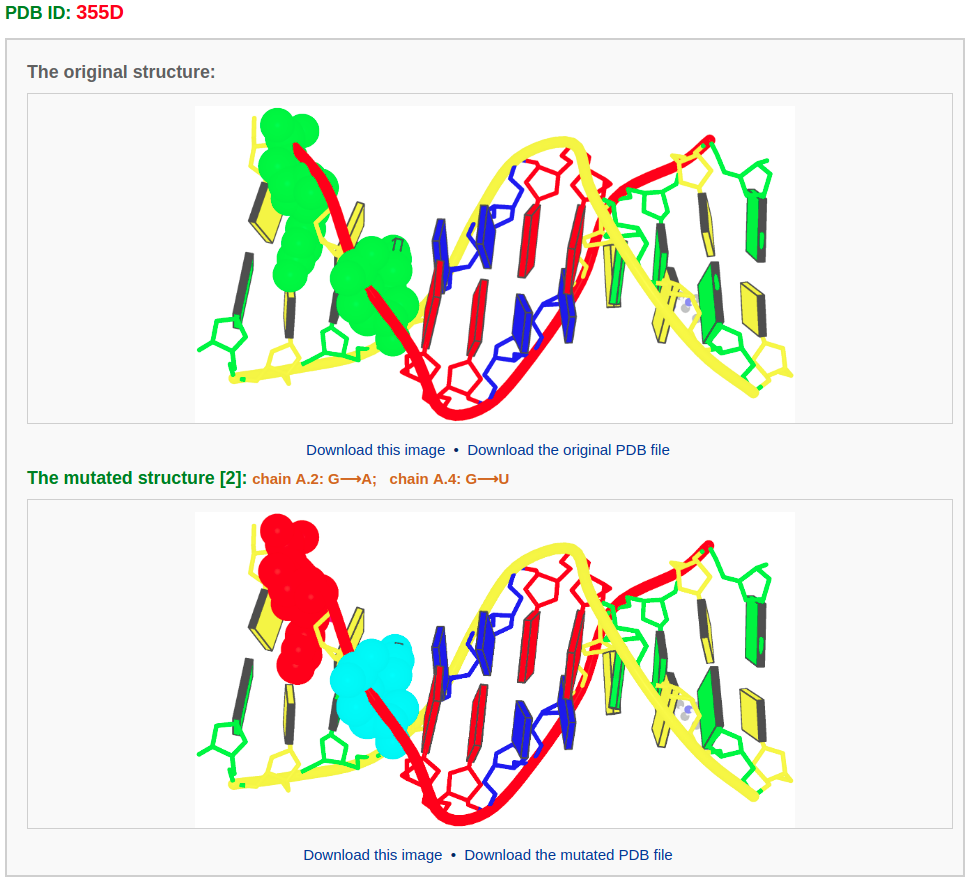
The text between the base-block images gives the number and identities of mutated nucleotides.
Example 6-2: Mutate a structure by uploading a PDB-formatted file

The user can also upload a PDB-formatted file to conduct base mutation(s). After uploading the structure and clicking the 'List all bases' button, the user follows a procedure that is similar to the example of mutating a structure by using the PDB/NDB ID.
Example 6-3: Construction of a G-quadruplex DNA model
The combination of mutation and fiber components can be used to build special DNA/RNA models, such as a G-quadruplex DNA model. The steps are as follows:
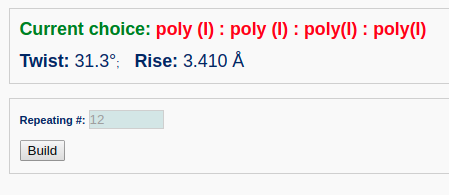
First, the user selects the poly(I):poly(I):poly(I):poly(I) fiber model from the list of "Commonly used fiber models".

For this non-generic fiber model, the user should enter the number of nucleotide repeating units, for example "12".
After clicking the ‘Build’ button, the user is directed to a results page containing the constructed model.
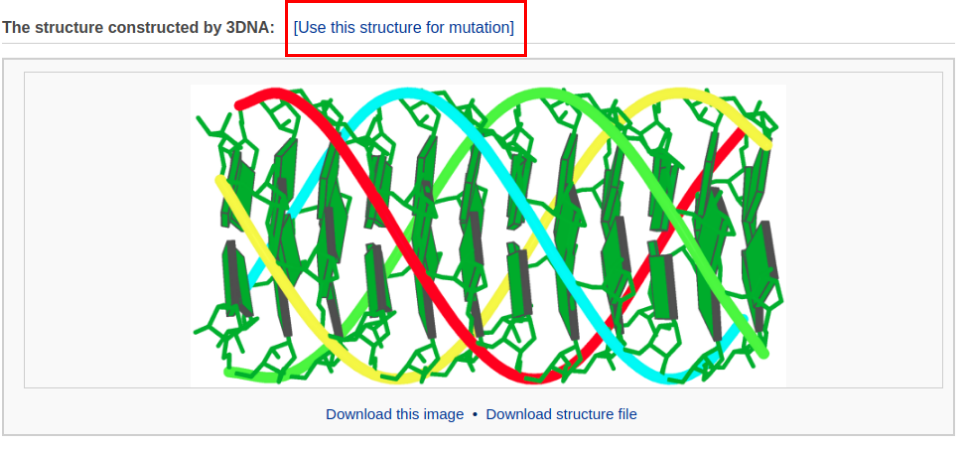

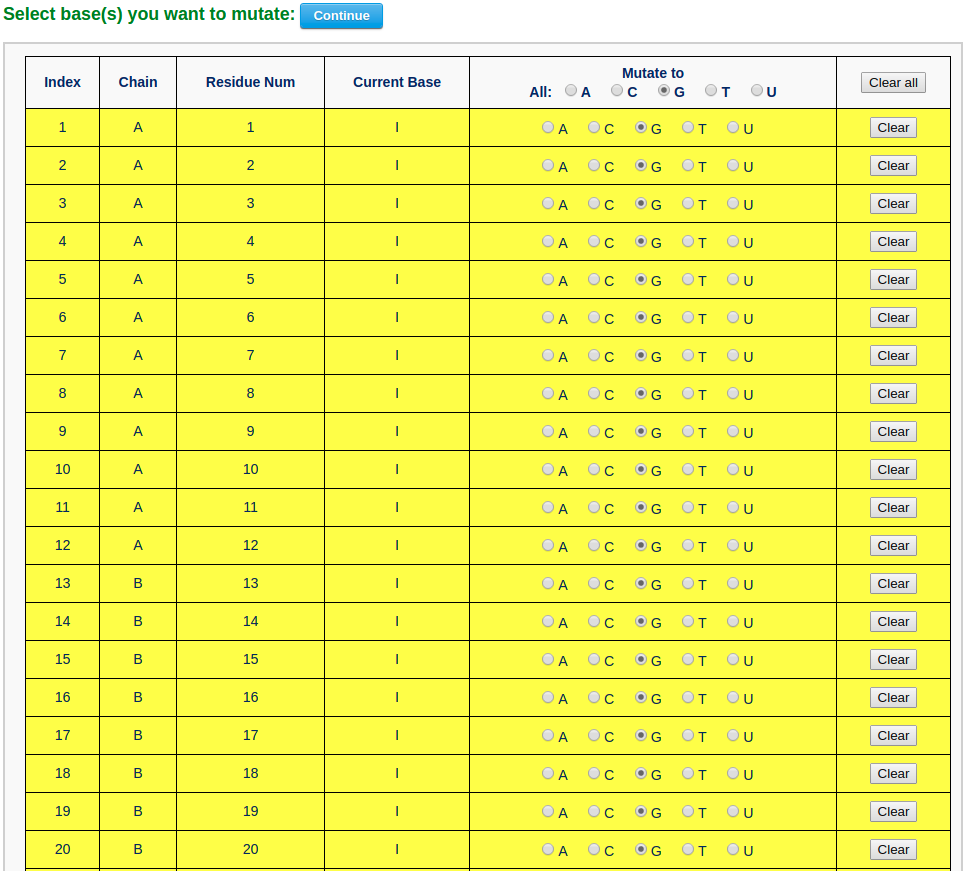
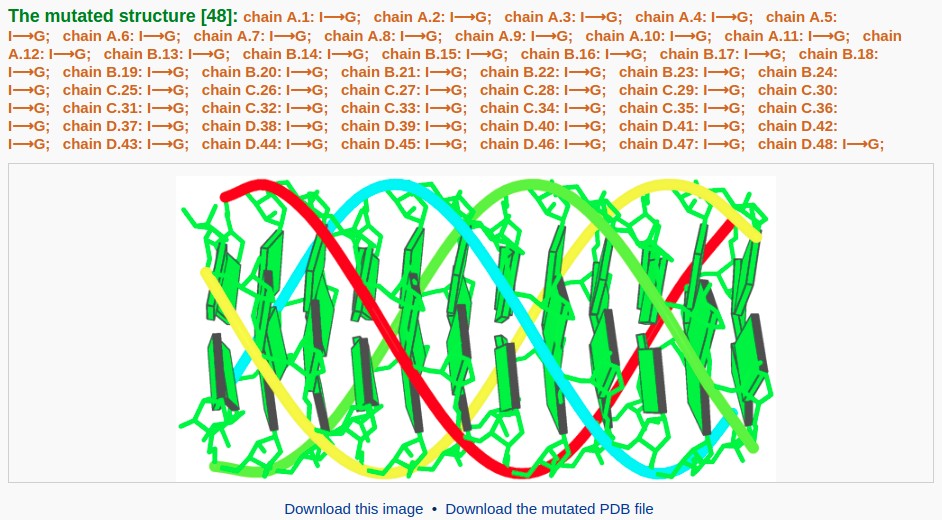
To build a G-quadruplex DNA model, the user needs to mutate all hypoxanthine bases in the current structure to guanine (G). Clicking the link "Use this structure for mutation" leads the user to a mutation selection interface. The interface contains a list of mutable bases. The user then chooses the option "Mutate to All G" and clicks the "Continue" button. The user will be directed to a results page with the mutated structure: a G-quadruplex DNA model.