|
Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures |
Tutorials
| Analysis | Visualization | Rebuilding | Composite | Fiber | Mutation |
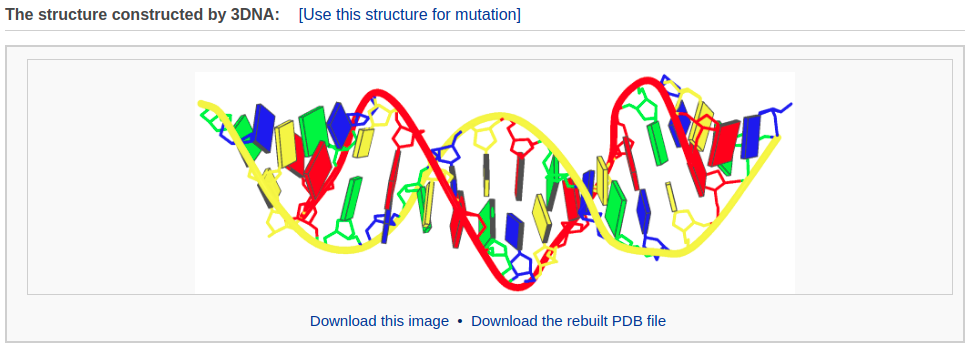
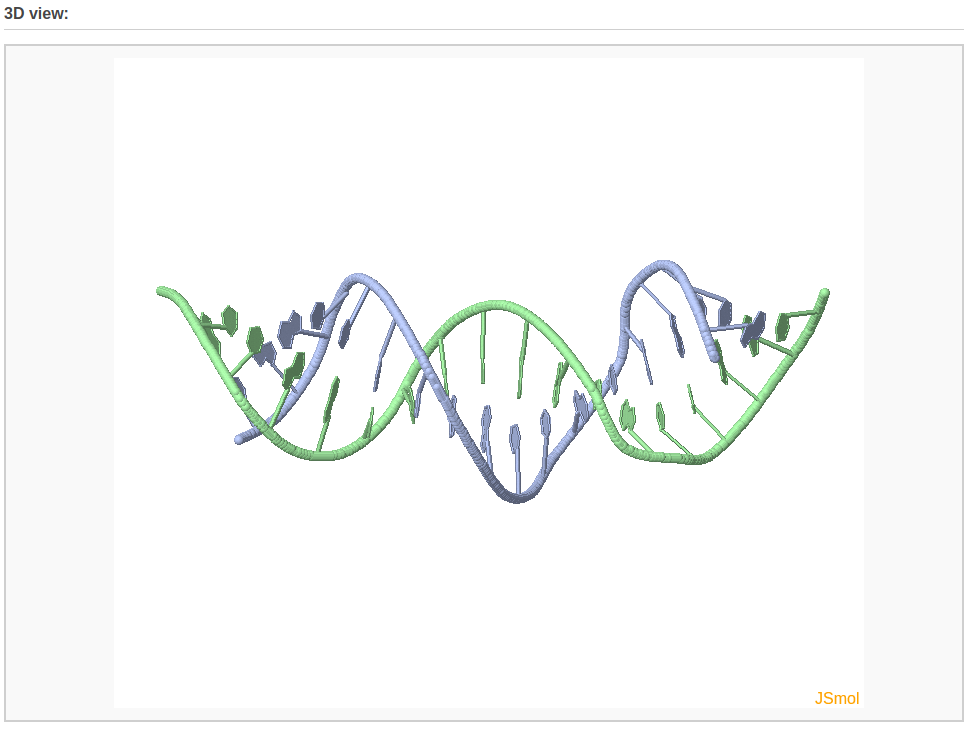
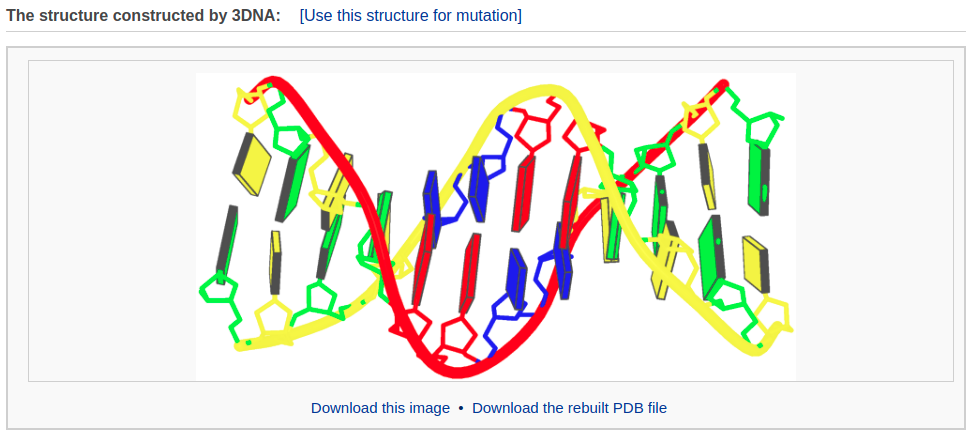
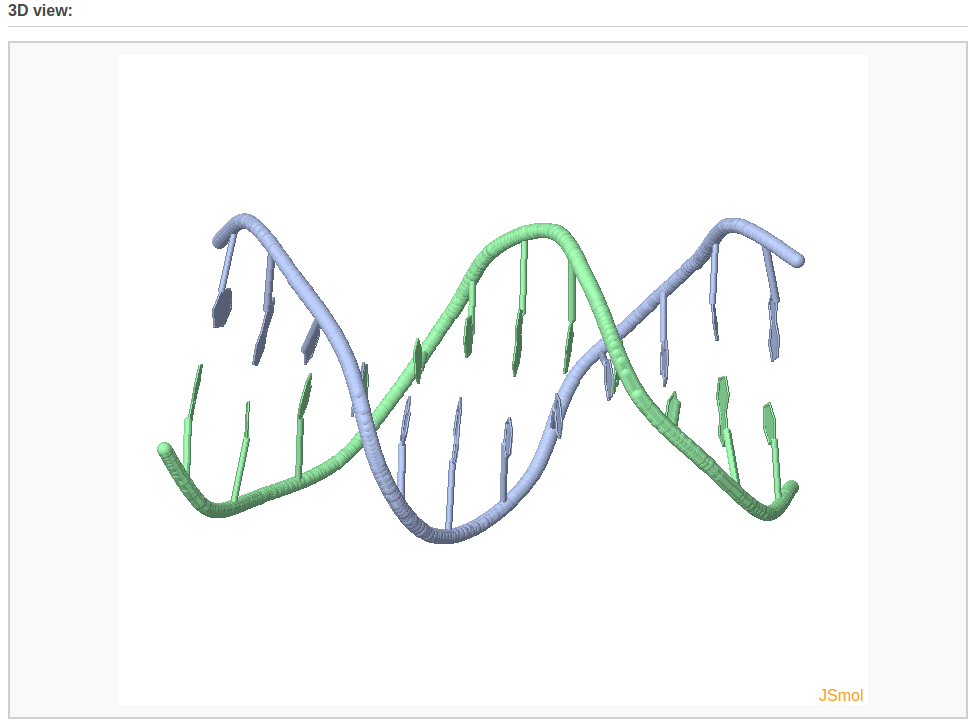
The rebuilding component of Web 3DNA 2.0 allows the user to build: 1) 3D models of customized double-helical and single-stranded structures with base pairs and base-pair/nucleotide steps arranged according to user-supplied rigid-body parameters, 2) a mixed DNA structure comprised of canonical A-, B-, and C-type DNA fragments. The output webpage includes a schematic representation of the constructed model, a PDB-formatted file with full atomic coordinates and a downloadable link, and a 3D interactive visualization of the constructed model.
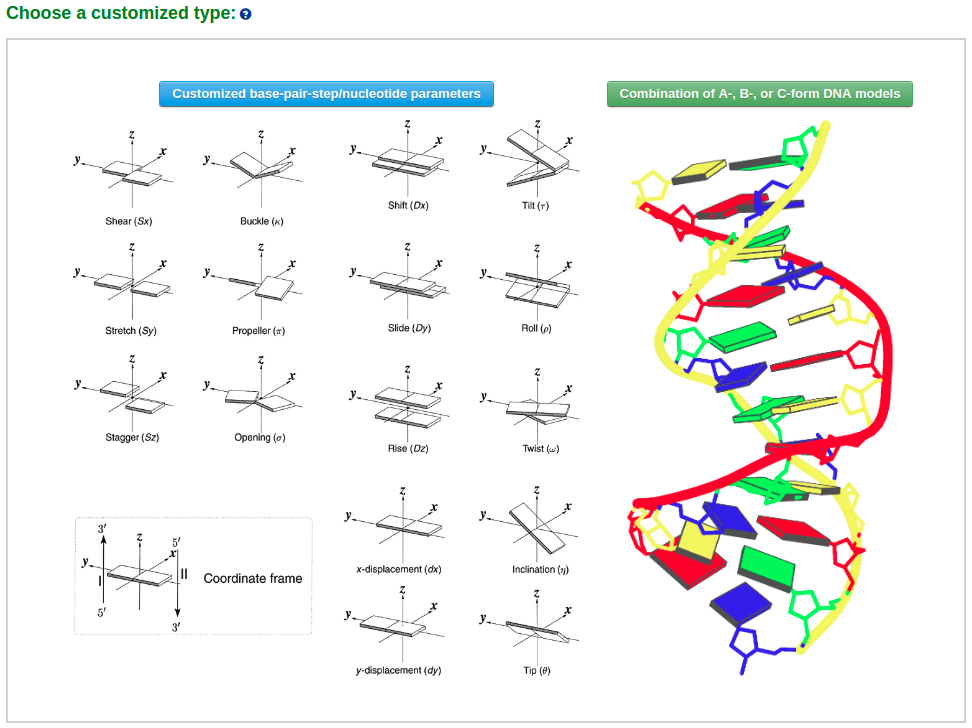
Example 3-1: Reconstruction of a customized model
The user must prepare a file containing the rigid-body parameters for the desired structure. Two example parameter files can be found at the "Test parameters" links. A single click of each button will automatically fill the corresponding content in the text area (one file for double-stranded DNA, the other one for single-stranded RNA). Note that the base-pair step parameters are null for the first base pair, but the parameter of the first base-pair can have non-zero values.
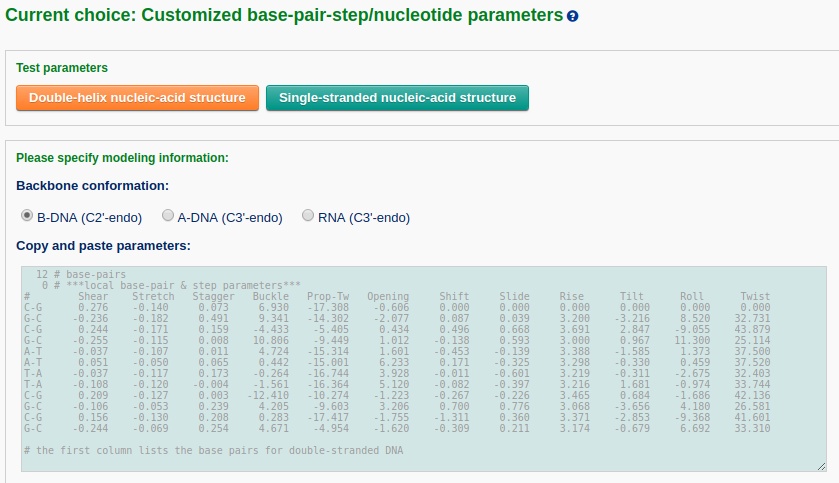
As an alternative, the user can also upload a parameter file in a valid format (please refer to the example parameter files: double-helix DNA and single-stranded RNA).

Next, the user needs to specify whether to construct a full atomic model and to select a sugar type: B-DNA (C2'-endo), A-DNA (C3'-endo), or RNA (C3'-endo). The user can also specify whether to optimize the backbone geometry.
After clicking the “Build Model” button, the user will be directed to a results page shown below.
Example 3-2: Construction of a model that combines of A-, B-, and C-DNA conformational segments

The user then can choose the desired number of segments in the model from a dropdown list (for example "3"). Clicking the "Next" button leads to an intermediate page where the user can select the sequence and conformational form (A-, B-, or C-DNA) of each segment. Note that the input sequence is that of the leading strand.
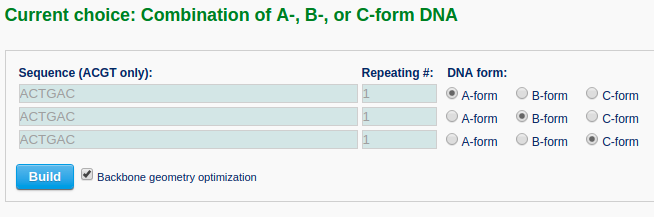
After clicking the ‘Build’ button, the user will be directed to a results page shown below.