|
Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures |
Tutorials
| Analysis | Visualization | Rebuilding | Composite | Fiber | Mutation |
The fiber component of Web 3DNA 2.0 allows the user to build fiber-diffraction models of nucleic acids with arbitrary sequence and helical types. A total of 56 fiber models is available in the current web server, with six commonly used ones (RNA, A-DNA, B-DNA, C-DNA, Pauling triplex, and poly(I) quadruplex models) placed at the top of the list.
After selecting a fiber model from the list, the user can specify a sequence and/or a repeating unit number. The output webpage includes a schematic representation of the constructed model, a PDB-formatted file with full atomic coordinates of base and sugar-phosphate atoms, and a 3D interactive visualization.
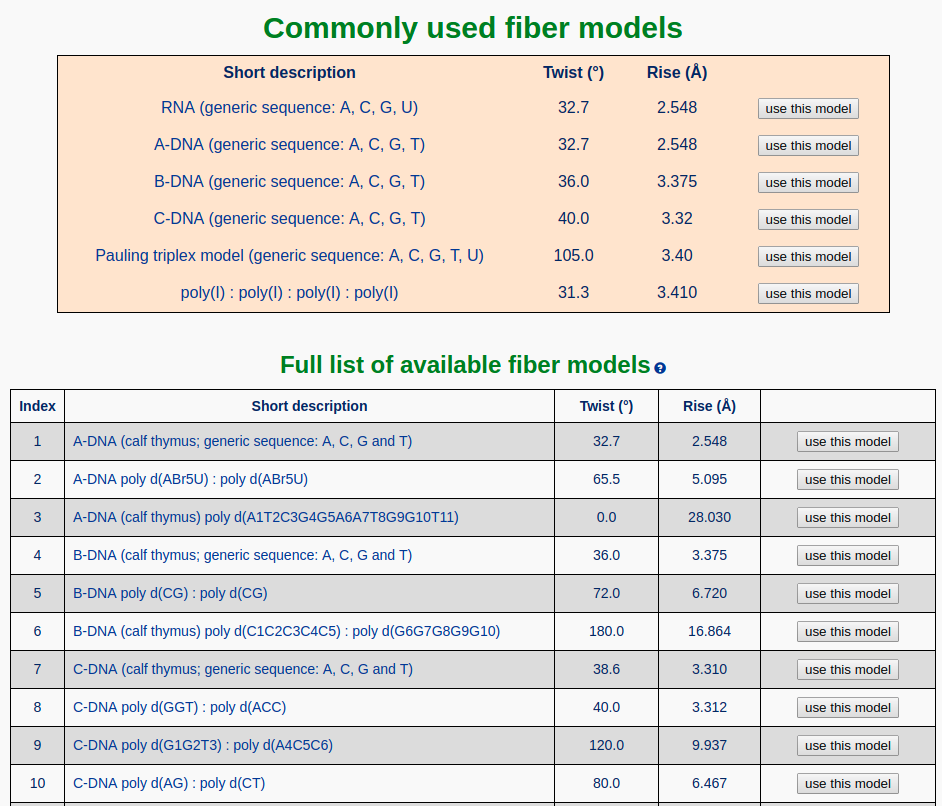
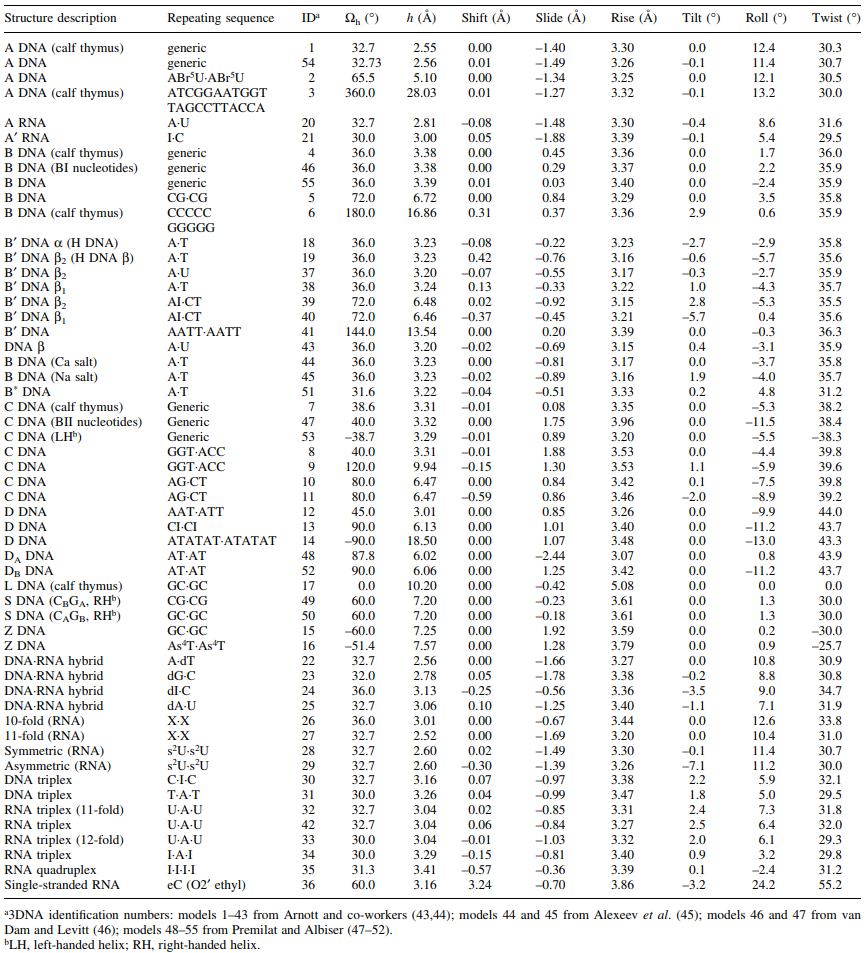
The detailed parameters of 55 fiber models are listed in the following table, taken from the initial 3DNA publication. For the Pauling triplex model, please check the blogpost "Pauling triplex model of nucleic acids is available in 3DNA" on the 3DNA homepage for further information.
Example 5-1: Construction of a generic B-form DNA fiber model
The user first selects the B-DNA (generic sequence: A, C, G, T) fiber model from the list of "Commonly used fiber models".

The generic DNA fiber models accommodate arbitrary base sequences of any length and only A, T, C, and G characters can be accepted in the input sequence. For a generic fiber, the user can type in a sequence and the number of repeating units as shown below. Note that the input sequence is that of the leading strand.
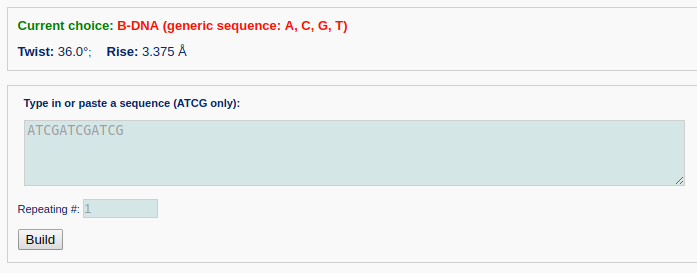
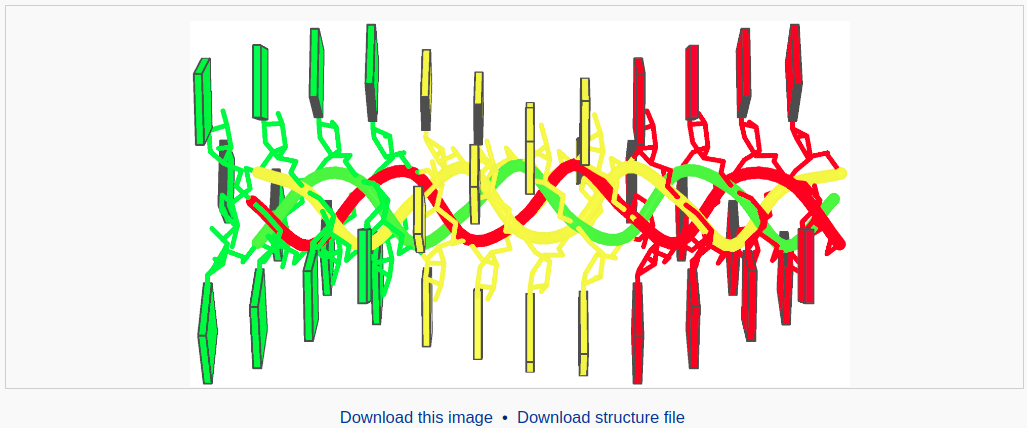
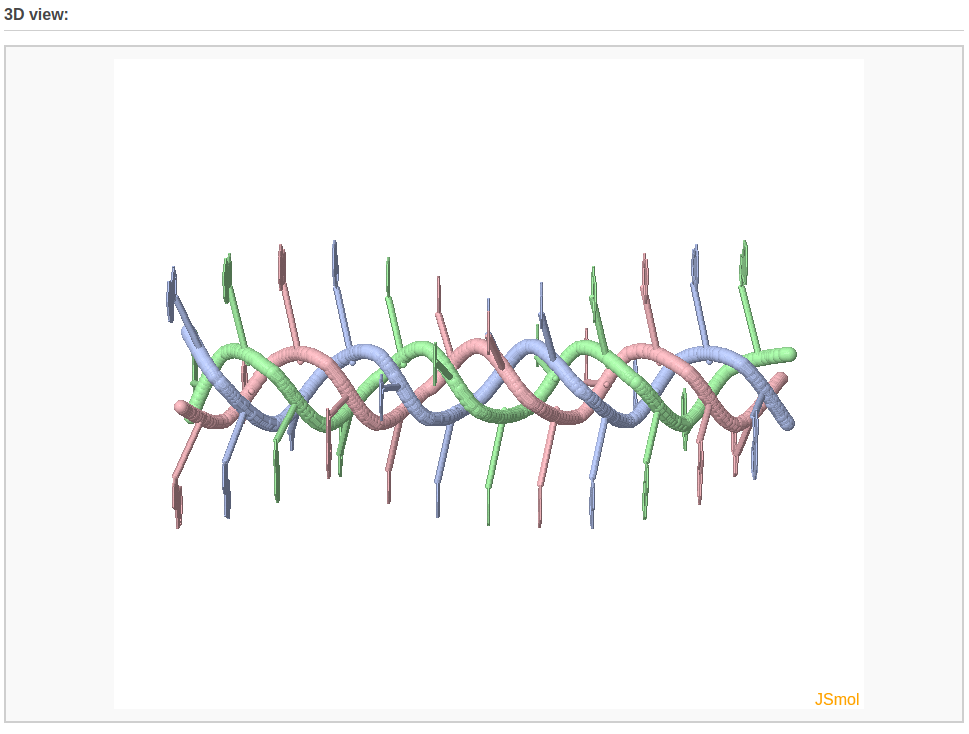
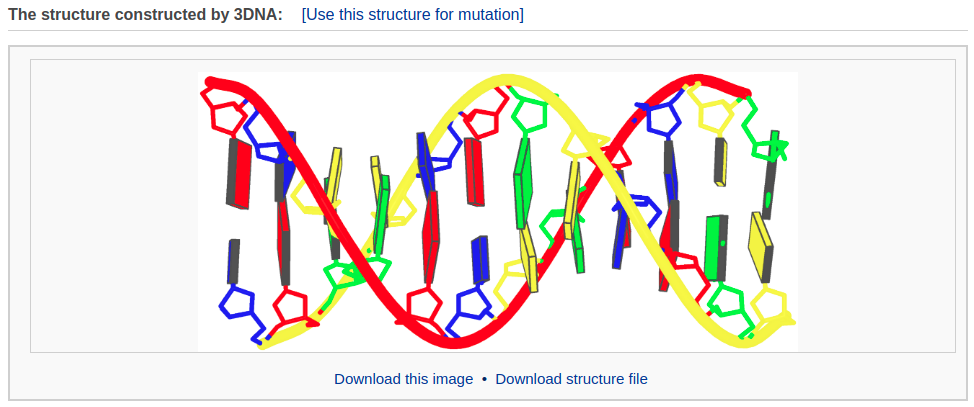
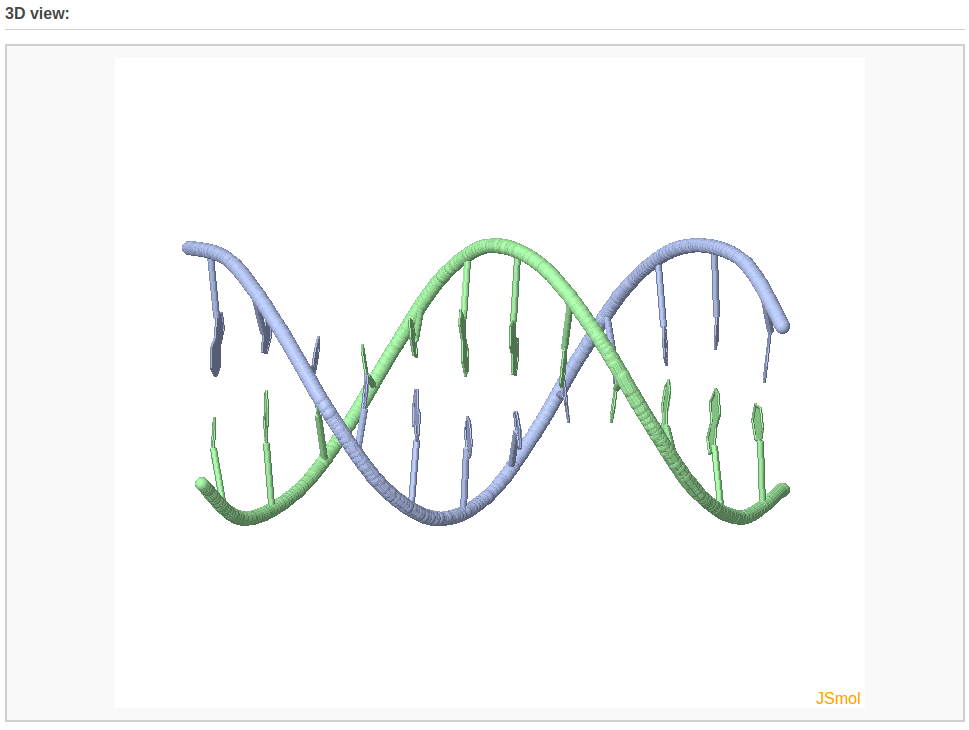
After clicking the ‘Build’ button, the user will be directed to a results page, which contains four sections: 1) a block representation of the constructed model, 2) a link to download a block image of the constructed model, 3) a link to download the coordinates of the constructed model, 4) a 3D interactive visualization of the constructed model.
Example 5-2: Construction of a Pauling triplex model
The fiber component also enables the user to build proposed DNA models. Here, we use the Pauling triplex model as an example.
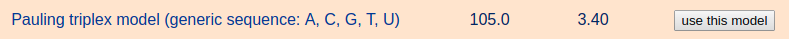
After selecting the Pauling triplex fiber model (generic sequence: A, C, G, T, U) from the list of "Commonly used fiber models", the user can enter a desired sequence, for example "AAAACCCCGGGG". The input characters are case-insensitive. Note that the input sequence is the same for all three strands.

After clicking the ‘Build’ button, the user is directed to a results page that contains four sections: 1) a block representation of the constructed model, 2) a link to download a block image of the constructed model, 3) a link to download the coordinates of the constructed model, 4) a 3D interactive visualization of the constructed model. The user should notice that the triplex model has phosphates in the center and bases on the outside of the structure.