|
Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures |
Tutorials
| Analysis | Visualization | Rebuilding | Composite | Fiber | Mutation |
The composite component of Web 3DNA 2.0 allows the user to construct models of DNA ‘decorated’ with proteins and other molecules by providing the requisite DNA-ligand structural templates and the locations of the binding sites. The output webpage includes a schematic representation of the constructed nucleic-acid protein/ligand complex, a PDB-formatted file with full atomic coordinates of the constructed model, and a 3D interactive visualization.
Example 4-1: Construction of a protein-bound DNA model
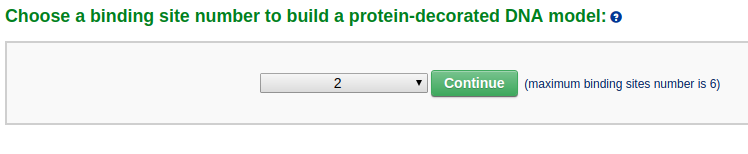
First the user needs to select the number of bound ligands from a dropdown list (for example "2"), and then clicks the 'Continue' button.
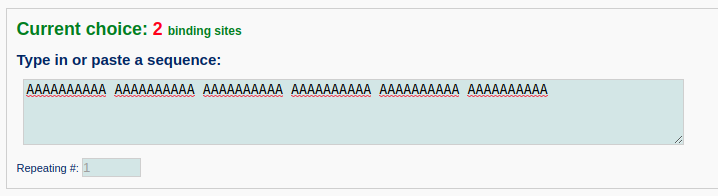
Next, the user is asked to type in the DNA sequence bound by the proteins of interest. Here, we enter a test DNA sequence of 60 base pairs with poly dA:poly dT. Note that the input sequence is that of the leading strand.
Then, the user needs to specify the conformation of the unbound DNA (A- or B-form) and to supply, for each binding site, the binding position. The binding position corresponds to the location of the central base or base-pair step of the specified protein-bound DNA segment. Here, two DNA-protein complexes (PDB ID: 1LMB and 5Z2T) serve as templates for binding DNA at positions 20 and 40.
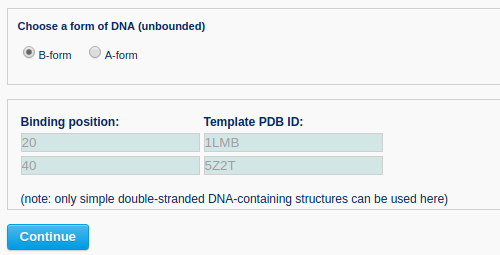
Clicking the ‘Continue’ button directs the user to a confirmation page, where the binding information can be verified. Warnings are given on this page if the binding input is inappropriate.
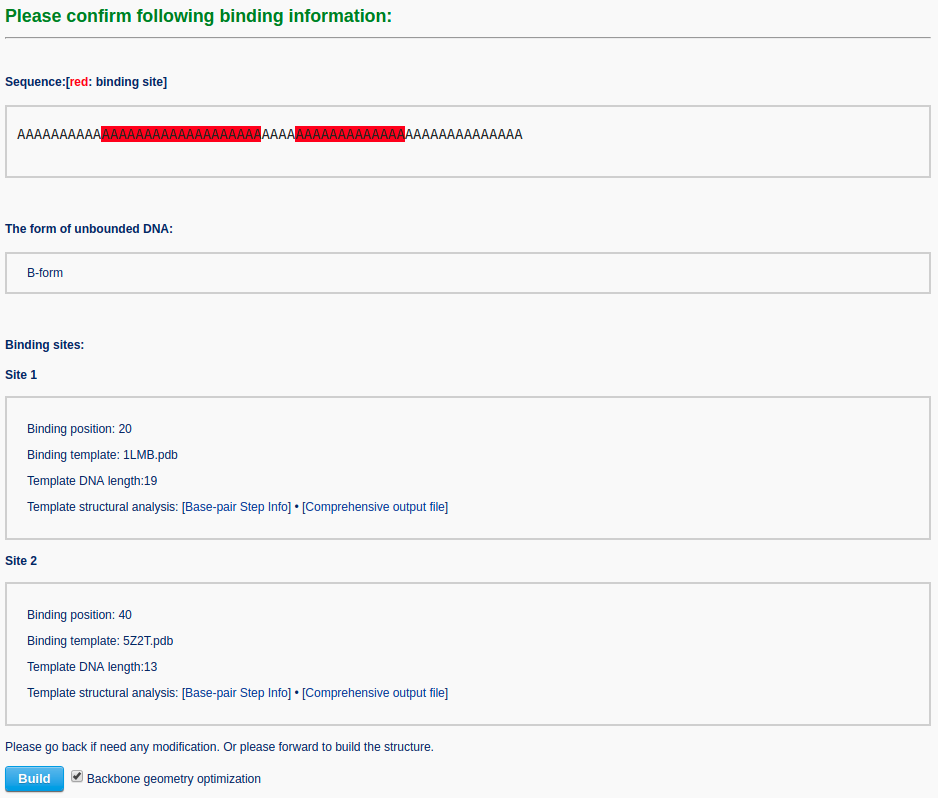
The user can choose whether to optimize the backbone geometry. After clicking the ‘Build’ button, the user will be directed to a results page that contains: 1) a ribbon-like representation of the DNA-protein complex, 2) downloadable links to the image and the atomic coordinate file, and 3) a 3D interactive visualization of the constructed DNA-protein complex.
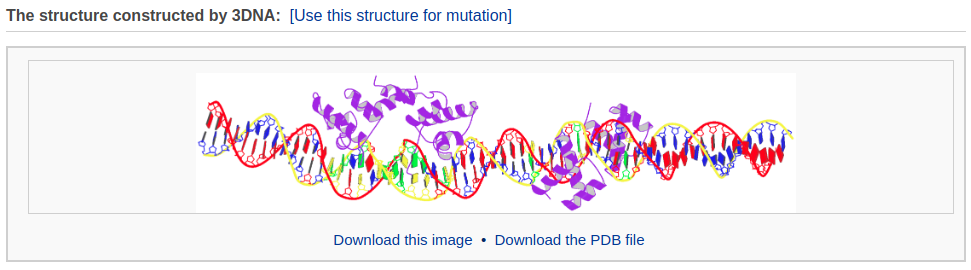
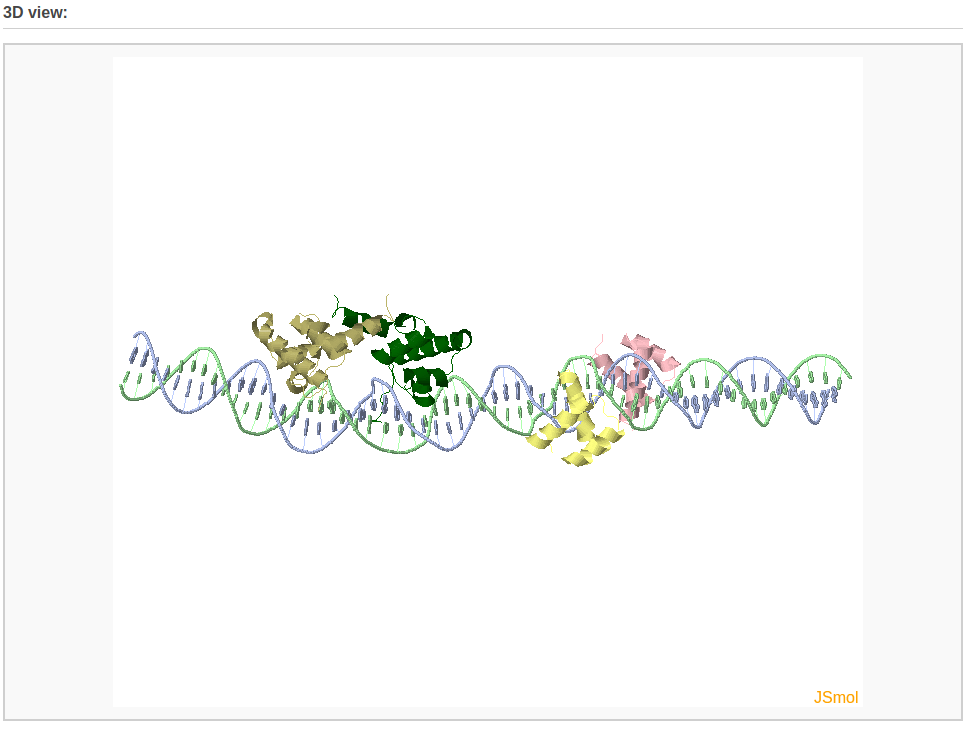
Example 4-2: Construction of a chromatin-like model
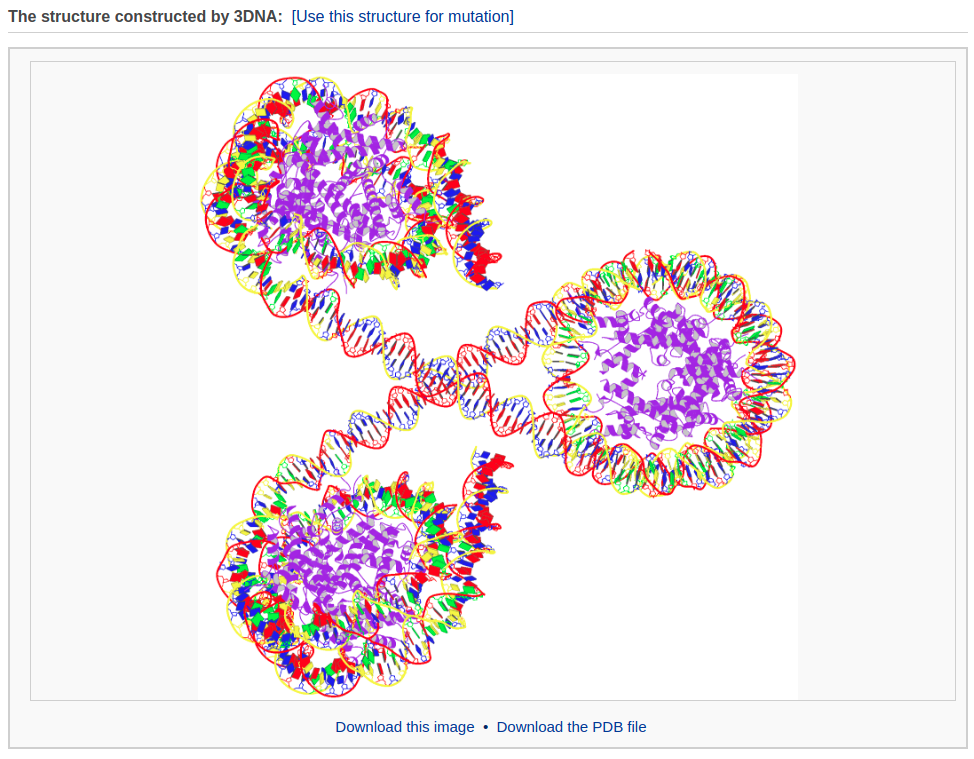
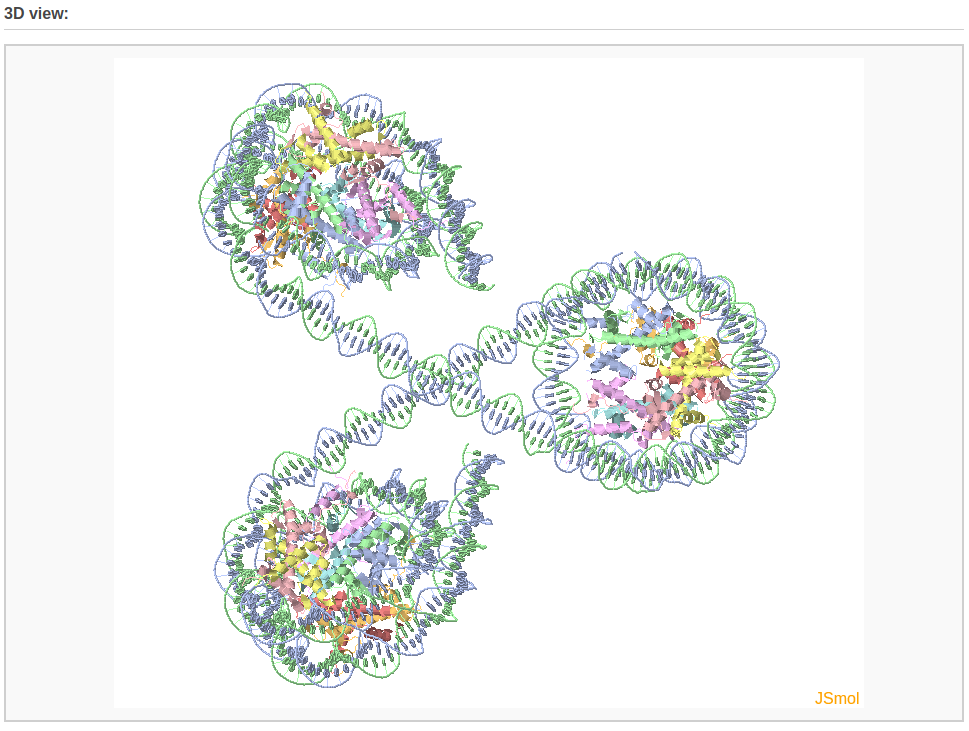
The composite component also enables the user to build complicated protein-decorated DNA structures, such as a chromatin-like model. Here, we use a three-nucleosome model as an example.
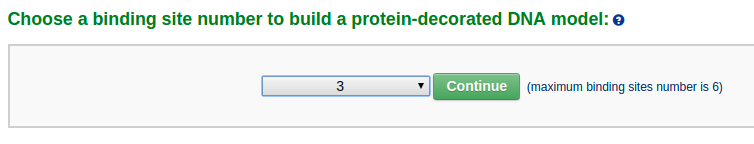
First the user assigns the number of nucleosomes in the final model to 3.
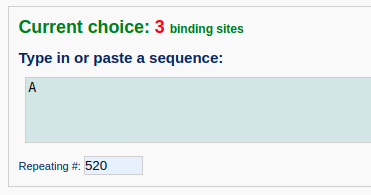
Next, the user enter a DNA sequence of poly dA:poly dT with the length of 520 base pairs. The user can just type in an "A" in the sequence text area and assign the repeating number to 520. Note that the input sequence is that of the leading strand.
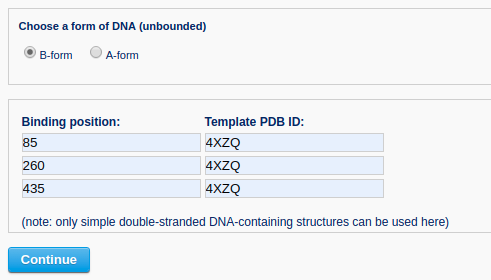
Then, the user specifies the conformation of the unbound DNA to B-form and provides nucleosome structures (for example PDB ID: 4XZQ) as templates to bind DNA at positions 85, 260 and 435.
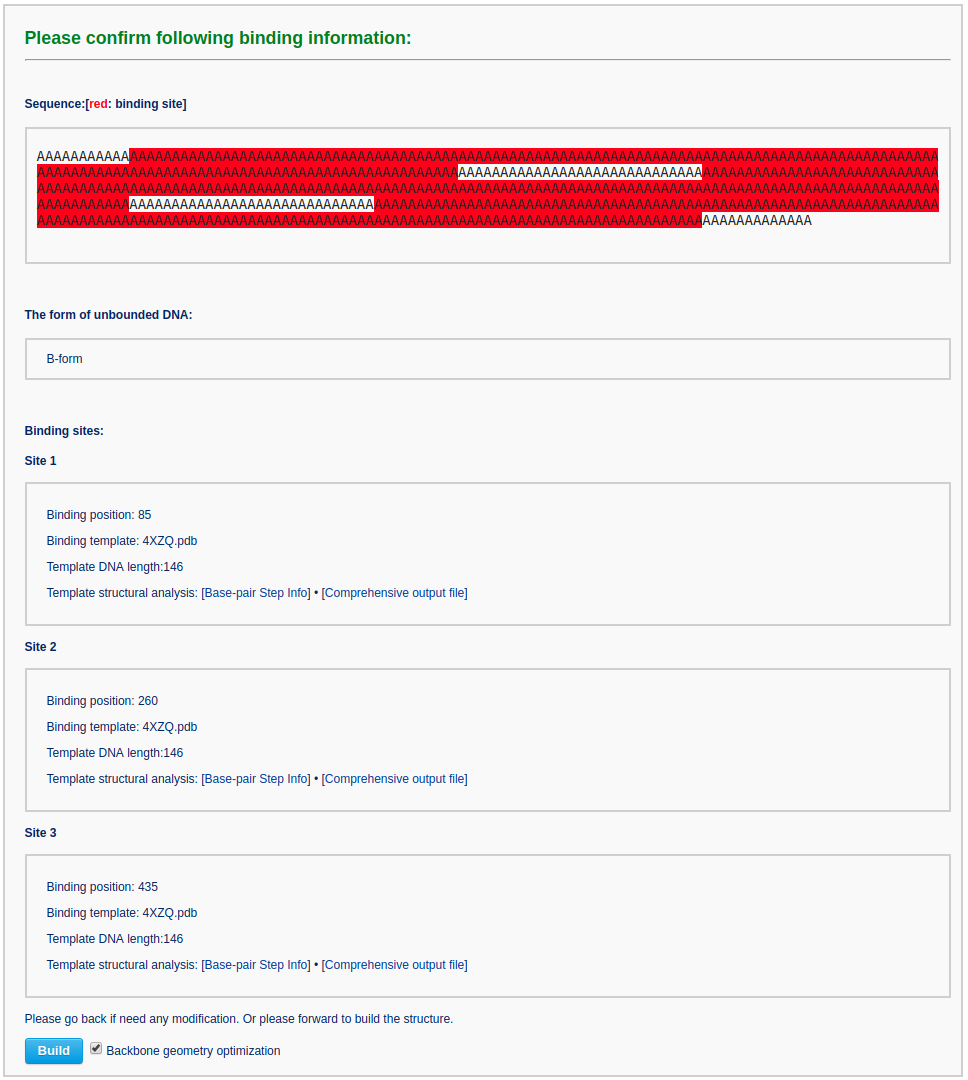
In the confirmation page, the user can verify all the binding information and then click the 'Build' button.
After waiting about 3 minutes, the user will obtain the constructed model: a chromatin-like structure.